Markov Chain Monte Carlo Example#
Inference of a distribution using Markov Chain Monte Carlo (MCMC) sampling (a very much simplified version of the implementation in BATEA). This a slightly more complicated implementation of what you saw in the MCMC rating curves. It is a simplified version of that found in BATEA (which is the hydrological model implementation that Brian Henn used in his work to determine what precipitation must have been given measurements of streamflow).
Note, this lab is designed to let you test the influence of selecting a gaussian or uniform prior, a gaussian or uniform liklihood, and of the number of samples needed, by comparing the MCMC approach to the theoretical distributions.
Derived from MCMCexample_noQN.m (Brian Henn, UW, February 2015)
import numpy as np
import scipy.stats as st
import matplotlib.pyplot as plt
%matplotlib inline
Inputs
# Specify prior distribution type ('uniform', or 'gaussian')
# this allows you to explore the impact of the shape of the prior distribution
PriorDistType = 'gaussian'
# Specify prior parameters (a=lower bound/mean, b=upper bound/st. dev)
aP = -2
bP = 2
# specify likelihood function type ('uniform', or 'gaussian')
LikelihoodDistType = 'gaussian'
# specify likelihood parameters (a = lower bound/mean, b = upper bound/st. dev.)
aL = 1
bL = 4
# MCMC parameters
nChains = 1
nSamples = int(1e5)
parJump = 1
dxMCMC = 1e-1
burn_in = int(1e2)
# Define evalBayes
def evalBayes(PriorDistType,LikelihoodDistType,aP,bP,aL,bL,x):
if PriorDistType == 'uniform': # For a uniform prior distribution
if (x >= aP) & (x <= bP):
xPrior = 1/(bP - aP)
else:
xPrior = 0
elif PriorDistType == 'gaussian': # For a Gaussian prior distribution
xPrior = st.norm.pdf(x,aP,bP)
if LikelihoodDistType == 'uniform': # For a uniform likelihood distribution
if (x >= aL) & (x <= bL):
xLikelihood = 1/(bL - aL)
else:
xLikelihood = 0
elif LikelihoodDistType == 'gaussian': # For a Gaussian likelihood distribution
xLikelihood = st.norm.pdf(x,aL,bL)
Post = xLikelihood*xPrior
return Post
Define “analytical” solution:
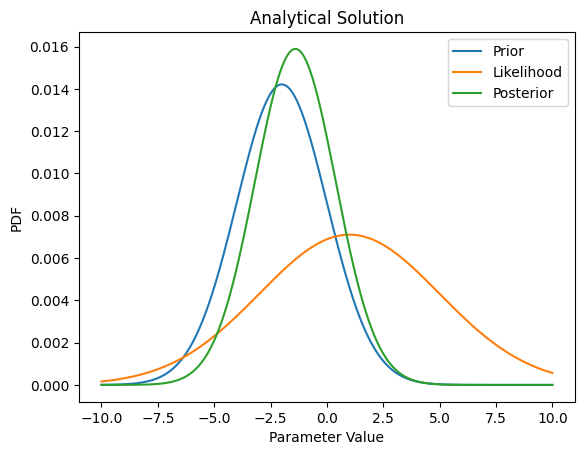
# define "analytical" solution
# Note that we are going to compare analytical updates to the prior (like we did in homework 5)
# with MCMC updates to the prior, as a way to test how well the MCMC method works.
X = np.linspace(-10,10,1001)
# Prior Distribution
if PriorDistType == 'uniform': # For a uniform prior distribution
PriorDist = np.zeros_like(X)
PriorDist[(X >= aP) & (X <= bP)] = 1/(bP-aP)
elif PriorDistType == 'gaussian': # For a Gaussian prior distribution
PriorDist = st.norm.pdf(X,aP,bP)
# Likelihood Distribution
if LikelihoodDistType == 'uniform': # For a uniform likelihood distribution
LikelihoodDist = np.zeros_like(X)
LikelihoodDist[(X >= aL) & (X <= bL)] = 1/(bL-aL)
elif LikelihoodDistType == 'gaussian': # For a Gaussian likelihood distribution
LikelihoodDist = st.norm.pdf(X,aL,bL)
PosteriorDist = np.zeros_like(X)
for i in range (0, X.size):
x = X[i]
PosteriorDist[i] = evalBayes(PriorDistType,LikelihoodDistType,aP,bP,aL,bL,x)
if np.sum(PosteriorDist) == 0:
scaleRatio = np.sum(LikelihoodDist)/np.sum(PriorDist)
else:
scaleRatio = np.sum(PosteriorDist)/np.sum(PriorDist)
plt.figure()
plt.plot(X,scaleRatio*PriorDist,label='Prior')
plt.plot(X,scaleRatio*LikelihoodDist,label='Likelihood')
plt.plot(X,PosteriorDist,label='Posterior')
plt.legend(loc='best')
plt.xlabel('Parameter Value')
plt.ylabel('PDF')
plt.title('Analytical Solution')
Text(0.5, 1.0, 'Analytical Solution')
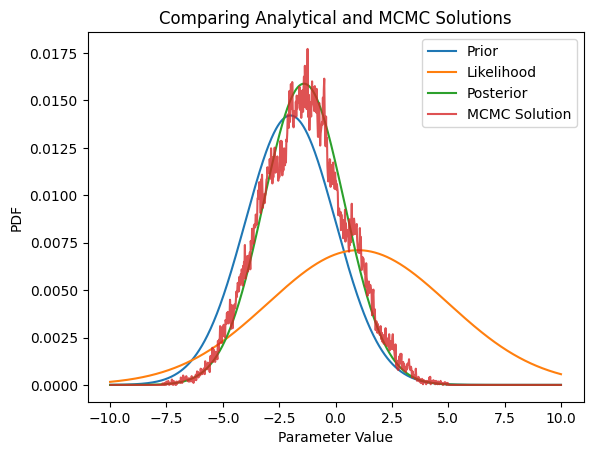
Now run the MCMC chains, using the most likely value of the prior as the starting point.
(Note that more clever methods exist to pick the starting point)
xStartMCMC = np.sum(X*PriorDist) / np.sum(PriorDist) #expected value, or mean of the pior distribution
fStartMCMC = evalBayes(PriorDistType,LikelihoodDistType,aP,bP,aL,bL,xStartMCMC)
Samples = np.empty([nSamples,nChains])
for i in range(0, nChains):
x = xStartMCMC
f = fStartMCMC
for j in range(1, nSamples):
# propose jump and decide
distJump = dxMCMC*np.random.normal()
fJump = evalBayes(PriorDistType,LikelihoodDistType,aP,bP,aL,bL,x + distJump)
if fJump >= f:
xNew = x + distJump
fNew = fJump
else:
randNum = np.random.uniform()
try: # try-except statement to handle zero division with uniform distributions
if randNum < (fJump/f)**parJump:
xNew = x + distJump
fNew = fJump
else:
xNew = x
fNew = f
except ZeroDivisionError:
xNew = x
fNew = f
x = xNew
f = fNew
Samples[j,i] = x
a = np.histogram(Samples[burn_in:],X)[0] * (np.sum(PosteriorDist)/Samples[burn_in:].size);
if np.sum(PosteriorDist) == 0:
scaleRatio = np.sum(LikelihoodDist)/np.sum(PriorDist)
else:
scaleRatio = np.sum(PosteriorDist)/np.sum(PriorDist)
plt.figure()
plt.plot(X,scaleRatio*PriorDist,label='Prior')
plt.plot(X,scaleRatio*LikelihoodDist,label='Likelihood')
plt.plot(X,PosteriorDist,label='Posterior')
plt.plot(X[1:],a,label='MCMC Solution',alpha=0.8)
plt.xlabel('Parameter Value')
plt.ylabel('PDF')
plt.title('Comparing Analytical and MCMC Solutions')
plt.legend(loc='best')
<matplotlib.legend.Legend at 0x7fcb62bc2d50>